Experimental Systems Course: RNA Interference
Course Lecturer: Prof Jamie Davies; e-mail Jamie.Davies@ed.ac.uk,
Tel (6)502999, lab web page http://golgi.ana.ed.ac.uk/Davieslab/
Please note: the most
up-to-date version of these notes will be found, while the course is
running, on http://golgi.ana.ed.ac.uk/coursenotes/
INDEX to
these course notes
General Introductory
remarks
(please read!)
Lecture
1 - Introduction to RNA interference
Lecture
2 - Using RNA interference as an experimental tool.
General
Introductory Remarks
RNA
inteference, until a couple of years ago a very obscure backwater of
molecular biological research, has suddenly developed into one of the
most talked about molecular biological techniques currently available.
Conferences are devoted to the subject, pharmaceutical companies are
throwing research money at it and the pages of Nature and Cell are packed with advertisements
trying to convince researchers that the companies that placed them are
each ahead of a very competitive game.
For this reason, the course team considers it essential that we cover
the essentials of RNA interference, even though the field is so new
that there is much still to learn and our best understanding is
expected to change quickly.
You will be able to find many excellent guides to RNAi on the web, but
be warned: I have been in this field for a few of years now, and one
of the most important things I find myself having to say to those
entering it is "be critical, and don't beleive the hype...". This is a
useful technique, but it is not an answer to all our prayers, however
much advertisers try to convey that impression..
1. The basic mechanism of RNAi
RNA interference (RNAi) refers to the ability of
double-stranded RNAs to shut down the expression of a messenger RNA
with which they have sequence in common.
RNAi was discovered independently in plants, fungi and
invertebrates, in response to attempts to genetically engineer these
organisms, and was at first given names such as 'gene silencing' and 'co-suppression'. History is probably not
the best way into the subject, because the earliest examples of RNAi
are not the best understood, so this lecture will jump straight to our
current understanding of RNAi and our best guesses about why the system
exists at all.
RNAi is triggered by double stranded RNA (dsRNA):
single stranded RNA (ssRNA), such as mRNA, cannot trigger it, although complementarty
ssRNAs can obviously come together in a cell to form a dsRNA that
can trigger RNAi. Cells have a number of protein complexes that
can recognize and bind to dsRNA, possibly because these proteins were
once part of a defence mechanism against RNA viruses (perhaps they
still are). The cells of vertebrates show a very powerful and (almost)
sequence-independent reaction to the presence of long dsRNAs, the
'interferon response', and this
complicates the story. In what follows, we will concentrate on
invertebrate cells and come back to vertebrates later.
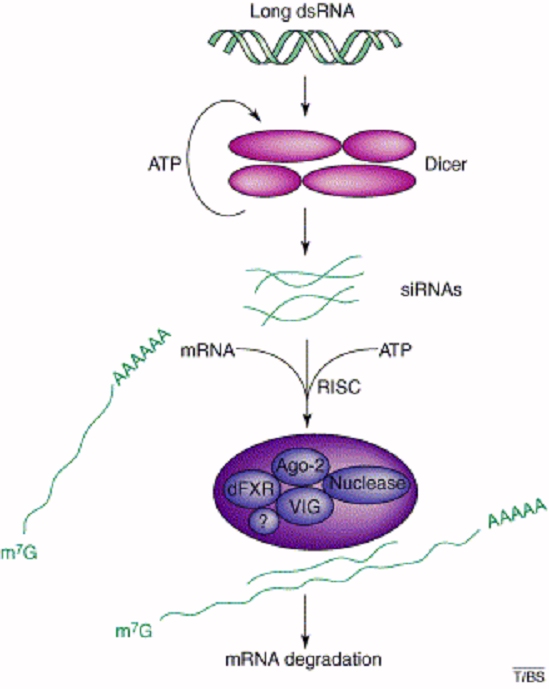
Long dsRNAs are recognized by a protein complex called Dicer, which cuts the dsRNA into short fragments of
about 22nt, this length being set by the structure of
the Dicer complex itself. An elegant experiment in which these short
degradation products were placed in Drosophila
cells in lieu of the long dsRNAs showed, to everyone's surprise at the
time, that these short lengths of RNA were capable of fully activating
the gene silencing response that the original dsRNA was. They are
therefore called siRNAs, for 'short interfering
RNAs'. They have overhanging ends (by 2 nt). When they are made by a
cell as a natural part of its genetic programme (rather than in
response to experimentally-introduced dsRNA), they are called microRNAs
(miRNAs - be careful not to misread this for mRNA).
The siRNAs associate with another complex of proteins, called RISC, which functions as an RNA-dependent RNA
endonuclease. Only siRNAs with the overhanging ends and 5' phosphates,
as produced by dicer, can recruit RISC. Once bound to siRNA, RISC is
activated and unwinds the siRNA to expose its single strands (or one of
its strands - this is not yet clear). If the exposed siRNA strand of a
siRNA-RISC complex happens to be complementary to part of an mRNA in
the cell, it will bind to this mRNA. On binding, RISC cuts up the mRNA,
thus preventing its expression.
Here is a cartoon of the basic mechanism of RNA, from TIBS:
RNAi is a catalytic process, because once the RISC complex has cut
up one mRNA, it is free to go off and attack another one, so that very
small amounts of siRNA can be used to clear a large amount of mRNA. In
principle, it is specific to mRNAs that are homologous to the dsRNA
sequences, and bystander RNAs are not affected (generally, this is true
- we will return to this point in lecture 2).
In animals such as C. elegans
and in plants, siRNA seems to be able to spread from cell to cell (in
the case of C elegans, via a
protein called SID-1), and tiny amounts of siRNA can silence the entire
organism, suggesting that amplification occurs.
In C. elegans, taregetting
the 3' region of a message with siRNA results in the generation of new
siRNAs for regions more 5'. This seems to happen because the animal has
an RNA-dependent RNA polymerase that can be primed by the siRNA/mRNA
hybrid. The newly synthesized dsRNA will be converted into siRNA by
dicer and so the cycle can repeat with more vigour. This also happens
in plants, but apparently not in vertebrates.
The effiiency by which RNAi works has been very useful for
experimenters (see next lecture), but it leaves the question about why
cells are so good at it. Recent discoveries of micro-RNAs that are
expressed during normal development raises the possibility thet RNAi is
a method of gene control, maybe a major method of gene control, in
plants and animals. There is, for example, evidence that they are used
even in a system as apparently well-understood as insulin production.
References
- Dicer - Zamore
PD,
Tuschl T, Sharp PA, Bartel DP.
- Interferon response - Sledz CA, Williams
BR
- That siRNAs can induce the full RNAi response - Elbashir SM, Lendeckel W,
Tuschl T
- RNA amplification - Sijen
T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH,
Fire A.
- Micro-RNAs in insulin production - Poy MN, Eliasson L,
Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T,
Rajewsky N, Rorsman P, Stoffel M.
- Evolutionary origin of endogenous interfering RNAs - http://www.nature.com/nature/journal/v453/n7196/full/453729a.html
- Endogenous micro RNAs and slicer (as an alternative to Dicer) - http://www.nature.com/nature/journal/v453/n7196/abs/nature07015.html
2. Using RNAi in
research
RNAi promises any researcher who can arrange to get dsRNA or siRNA into
the correct cell at the correct time a way of knocking out (or at
least, down) any gene she wishes to target.. To understand why this is
so important, it is important to consider why we want to suppress the
action of genes, and the limitations of alternative ways of doing this.
Deleting the expression of a gene has, for a long time, been the most
powerful way of testing a hypothesis about the function of that gene.
If ruddyconk2 is thought to
be the gene that causes reindeer to develop red noses, then knocking
out the gene and observing the colour of the noses produced is a
powerful way of testing whether this is true (if the nose is still red,
ruddyconk2 is not required for
making a red nose; if it is not red, then the gene is needed).
There are several methods for removing the function of a gene, all with
advantages and disadvantages;
- Transgenic knockout: this has the advantage of being absolute,
but the disadvanatge that if ruddyconk2
is also needed to make, say, a placenta, then the embryo will not
survive for long enough for a researcher to see if it has a red nose.
- Conditional knockout (eg cre-lox): this can arrange for a
knockout to work only in some tissues - it is great when drivers are
available that are expressed onyl in the tissue of interest (for early
development, esepcailly, this is often a big problem) but it is
expensive and takes a coupld of years typically.
- Pharmacological inhibition: this is useful in that a drug can be
applied at any time, and therefore you can let something develop
normally to stage X and then do the knockout, but has the problem that
very few proteins are specifically inhibited by drugs, althoug the list
keeps growing.
- Antibodies etc: These are fine for inhibiting extracellular
proteins (eg growth factors), but are useless for getting inside the
cell.
The use of RNAi promises all of the advantages of
pharmacology, but with absolutely any protein being targetted, and in
principle experiments could be done in mere weeks without requiring
animal breeding etc.
In this lecture, I shall concentrate on applying RNAi
to mamalian systems, because this is where most effort is being
expended (because RNAi may prove a very valuable anti-viral and
anti-cancer treatment, as well as being a research tool, and because
mammals do not have the clever genetic tricks available in worms and
flies so new techniques are especially valuable).
Because they have the interferon
response, mammalian cells cannot be treated with long dsRNAs, so have to be treated with siRNAs
instead. There are several ways of doing this;
- Synthesize siRNAs chemically, and apply to cells in some suitable
membrane-crossing complex
- Make siRNAs from a plasmid in vitro, then treat as above
- Transfect cells with a plasmid that makes siRNA
- Transfect cells with an inducible plasmid that makes siRNA.
Designing siRNAs is not straightforward, although there are some useful
guidelines. Generally, only about 1/4 work well, and people identify
these by simple tests in cell lines before going on to do anything
complicated.
All siRNA experiments require good controls. These are, typically;
- Negative control cells with no siRNA
- Negative cells with an irrelvant siRNA
- The use of several different siRNAs to the same target, to
control for unexpected 'friendly fire' incidents on other messages
(off-target effects).
- Careful monitoring of protein knockdown (time, extent, % of cells)
- Careful monitoring that other proteins are not knocked down.
- If possible, rescue of the cells with a plasmid endocing the same
protein but using a different DNA sequence (codon redundancy).
Careful use of controls have demonstrated that there are several
surprising effects of dsRNAs that do not act via the pathway outlined
in lecture 1. One is induction of the interferon pathway. Another is
activation of TLR3 on the outside
of cells; this also induces an interferon/IL12 type response and blocks
growth of blood vessels, something originally thought to operate via
the specific RNAi pathway.
References for lecture 2
- Use of siRNA in mammalian cells - Elbashir SM, Harborth J,
Lendeckel W, Yalcin A, Weber K, Tuschl T.
- Use of siRNA to test later functions of a gene - Davies JA, Ladomery M,
Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N.
- Good descriptions of typical RNAi technology - http://www.ambion.com/techlib/resources/siRNA/index.html
- The TLR3 path - http://www.nature.com/nature/journal/v452/n7187/full/452543a.html
(this is a news and views review with refs to the original paper).
- dsRNAs as activators of
transcription - http://www.nature.com/nature/journal/v448/n7156/full/448855a.html
- I am not going to cover this in the lectures but if you take the
trouble to read it you could be very impressive in a variety of exam
questions!
- Off-target effects - statistics - http://www.nature.com/nature/journal/v443/n7109/abs/nature05179.html
- I am not going to cover this in the lectures but if you take the
trouble to read it you could be very impressive in a variety of exam
questions!
Reference List
These references serve two purposes, to help anyone I might have
confused,by
repeating the lecture material in someone else's words (where
possible),
and to allow everyone to explore the topics in more detail. This
material is too new to be in textbooks, but I am aware that you do not
have time, with the exams so close, to spend trawling through original
papers. If you want more detail, use these references, but if you just
want to cram for the exam use your lecture notes and websites such as
Ambion's for quick summaries.
Davies
JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N.
(2004) Development of an siRNA-based
method for repressing
specific genes in renal organ culture and its use to show that the Wt1
tumour suppressor is required for nephron differentiation. Hum
Mol Genet. 2004 Jan 15;13(2):235-46 PMID: 14645201
Elbashir
SM, Lendeckel W, Tuschl T (2001) RNA
interference is mediated by 21- and 22-nucleotide RNAs. Genes
Dev. 2001 Jan 15;15(2):188-200. PMID: 11157775
Elbashir
SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in
cultured mammalian cells. Nature. 2001 May
24;411(6836):494-8. PMID: 11373684
Poy
MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer
S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. (2004) A pancreatic islet-specific microRNA regulates insulin
secretion. Nature. 2004 Nov 11;432(7014):226-30. PMID: 15538371
Sledz
CA, Williams BR (2004) RNA interference and
double-stranded-RNA-activated pathways. Biochem Soc
Trans. 2004 Dec;32(Pt 6):952-6. PMID 15506933
Sijen
T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH,
Fire A. (2001) On the role of RNA
amplification in dsRNA-triggered gene silencing. Cell. 2001 Nov
16;107(4):465-76 PMID: 11719187
Zamore
PD, Tuschl T, Sharp PA, Bartel DP (2000). RNAi:
double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to
23 nucleotide intervals. Cell. 2000 Mar 31;101(1):25-33. PMID
10778853
Glossary
Co-suppression - another synonym for RNAi,
usually used by plant and fungi folk.
Dicer - the
protein complex that binds to long dsRNAs and
chops them to siRNAs
dsRNA - double stranded RNA
Gene silencing - synonym for RNAi.
Interferon
response - a slightly slang phrase (in that it is used quite
loosely) for the non-sequence-specific response of mammalian cells to
long (>20bp or so) dsRNAs.
mRNA - messenger
RNA
nt - nucleotide (a
way of counting length that is equally useful for ssRNA
and dsRNA).
RISC - the
nuclease complex that assmebles around siRNAs and cuts target RNAs
complementary to them.
RNAi - RNA
interference: this is the general term for the ability of
double-stranded RNAs to block the expression of a messenger RNA with
which they have a sequence in common (or almost in common).
siRNA -
short interfering RNA - the product of dicer cutting of long dsRNA. siRNA is sometimes chemically synthesized for
application to cells.
ssRNA -
single stranded RNA
Date of this file: 5th November 2012. Jamie.Davies@ed.ac.uk

